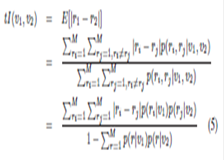Biomedical Image & Data Analysis Lab - BIDAL
We are interested in developing an intuitive computational framework to better analyze medical and biological data in order to innovate the way such data are used in the current clinical practice and scientific research. Our research explores various computational theories for developing new robust, efficient and/or large-scale algorithms and seeks their practical applications to solve clinically and scientifically significant problems, such as cancer prevention. BIDAL is located in San Francisco State University and affiliated with SFSU’s Department of Computer Science.
News:
Prof. Okada presented Keynote Talk at College of Science and Engineering Showcase Event, San Francisco State University, May 2017
Professors team up to design a better prosthetic arm, SF State News, Mar 2016
Computer technology aids medical advances, SF State News, Feb 2015
|
|
Kazunori Okada Principal Investigator, Professor, CS Dept. SFSU Ph.D. in Computer Science at University of Southern California |
|
|
Yoshimasa Iwano (MSc Student, CS Dept, SFSU: Aug/2022 – Current) Project: Computer Vision Applications for Ocean Fishery Management Links: yiwano AT sfsu.edu |
|
|
Chris Huber (MSc Student, CS Dept, SFSU: Jan/2023 – Current) Project: Covid-19 Diagnosis with 3D CT Scans Links: chrish AT mail.sfsu.edu |
|
|
Ekarat Buddharuksa (MSc Student, CS Dept, SFSU: Aug/2023 – Current) Project: Object Tracking for Underwater Fish Detection Links: ebuddharuksa AT sfsu.edu |
|
|
Marie Shimizu (MSc Student, CS Dept, SFSU: Aug/2023 – Current) Project: Multimodal Stress Level Assessment Links: mshimizu4 AT sfsu.edu |
|
|
Aaron Huber (BS Student, CS Dept, SFSU: Aug/2023 – Current) Project: Multimodal Stress Level Assessment Links: akuo6 AT sfsu.edu |
|
Alumni |
Xinwei Fan (MSc, May 2022) currently at Amazon Web Services Jakob Dohrmann (MSc, Dec 2020) currently at Cisco Katie Fotion (MSc, May 2018) currently at Cruise Jeff Hung (MSc, May 2018) currently at Genentech, Inc Saman Porhemmat (BSc, Feb 2018) currently at UC Irvine Maya Stark (MSc, Feb 2018) currently at Genentech, Inc Juniper Overbeck (BSc MATH, Dec 2017) currently at SFSU Andre Simpelo (BSc, May 2017) currently at Udacity Octavian Drulea (MSc, May 2017) currently at Driver Juris Puchin (MSc, May 2017) currently at Acuity Brands Rupal Khilari (MSc, May 2017) currently at The Mill Serge Dusheyko (MSc EECS, May 2017) currently at Colorado Electronic Product Design, Inc Diana Chu (BEng, Dec 2016) currently at Cornell University Poulomi Das (BSc, May 2016) currently at UCSF Medical Center Nao Funada (BSc, May 2015) currently at Softbank Corp Marzieh Golbaz (MSc, May 2015) currently at Evidera Sergey Sergeev (MSc, May 2013) currently at SquareTrade John Collins (MSc, May 2013) currently at Oregon Child Development Coalition Oliver Newland (MSc, May 2013) currently at CrowdFlower Eric Chiang (BS, Jan 2013) currently at CoreOS Hong Ge (Visiting Professor: Jan 2012- Jan 2013) South China Normal University, China Malik Fahem (Visiting Student: Jul 2012 – Aug 2012) ENSEEIHT, Toulouse, France Jing Huo (PhD, 2012) GBM Tumor Analysis, UCLA, currently at TeraRecon Inc Asha Rani Hongenalli Chandrappa (MSc, 2012) RTI Project, currently at Assurant Miyuki Suzuki (MSc, Dec 2011) currently at Capital One Runtang Wang (MSc, Feb 2011) currently at The Mind Research Network Niranjan Timilsina (MSc, Sep 2010) currently at Trulia Yang Zhao (MSc, May 2010) currently at GoDaddy Andrew Scott (BS, May 2010) currently at SFSU Ngoc Lam-Miller (MSc, Oct 2009) currently at EMC/Greenplum Xinhang Shao (MSc, Oct 2009) currently at Silicon Valley Education Foundation Mehran Kafai (MSc, May 2009) currently at A9.com Yiyi Miao (MSc, Feb 2009) currently at OPSWAT Rodrigo Salas (MSc, Feb 2009) currently at Intel Corporation David Wang (BS, Dec 2008) currently at CSU East Bay Edgar Liu (MSc, May 2008) currently at Acer Arturo Flores (MSc, May 2008) currently at KAB Laboratories Steven Rysavy (MSc, May 2008) currently at Pacific Northwest National Laboratory Sean Todd (BS, May 2008) currently at Winasaurus Ryan West (BS, Aug 2007) currently at Location Labs |
Selected current and previous projects are summarized in the below table.
|
|
Integrating Facial and Speech Recognition for Stress Level Assessment in College Students: Online stress level assessments, particularly beneficial for college students as an inexpensive and accessible resource, offer broad prevention for stress-related mental health crises. Stress detection has been extensively researched in related fields using various markers. Concerning online applications, significant emphasis has been placed on user activities and text data, neglecting subtle discrepancies in facial and vocal data. We propose an approach that enhances accuracy by offering personalized stress detection through integrating ques from multiple channels including text, vocal, and facial markers. |
|
|
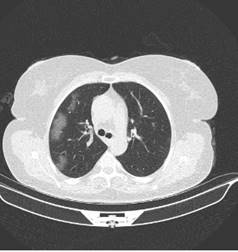
Covid-19 Detection from lung CT Scans using DNN with limited resources: Effectiveness of using Deep Neural Network (DNN) for detecting and diagnosing covid-19 infections in lung CT scans have been demonstrated in the literature, however building such model to process 3D images such as CT scans demands availability of high-power computational resources whose accessibility maybe limited especially in developing nations. This research investigates methodologies for transfer learning that builds a 3D model from 2D models, exploiting existing well-performing pretrained models designed to classify 2D images while minimizing the computational demands for building 3D models. |
|
|
Computer Vision Applications for Ocean Fishery Management: Ocean fishery plays an important role in addressing food shortage worldwide however understanding biology of fish and its largescale ecosystems remains largely uncharted due to lack of means to observe fish in their habitat continuously and quantitatively. This research aims to develop tools to analyze fish behaviors in situ by adapting computer vision solutions: visual detection, segmentation, and object tracking. |
|
|
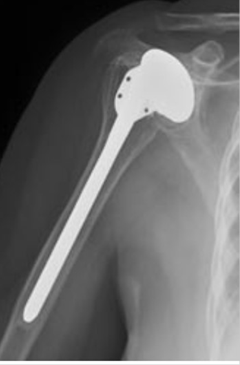
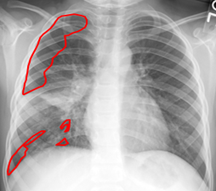
Shoulder Implant Classification in X-Ray Images: Total Shoulder Arthroplasty (TSA) is a type of surgery in which the damaged ball of the shoulder is replaced with a prosthesis. Many years later, this prosthesis may be in need of servicing or replacement. In some situations, such as when the patient has changed his country of residence, the model and the manufacturer of the prosthesis may be unknown to the patient and primary doctor. Correct identification of the implant's model prior to surgery is required for selecting the correct equipment and procedure. We present a novel way to automatically classify shoulder implants in X-ray images. We employ deep learning models and compare their performance to alternative classifiers, such as random forests and gradient boosting. We find that deep convolutional neural networks outperform other classifiers significantly if and only if out-of-domain data such as ImageNet is used to pre-train the models. In a data set containing X-ray images of shoulder implants from 4 manufacturers and 16 different models, deep learning is able to identify the correct manufacturer with an accuracy of approximately 80% in 10-fold cross validation, while other classifiers achieve an accuracy of 56% or less. We believe that this approach will be a useful tool in clinical practice, and is likely applicable to other kinds of prostheses. |
|
|
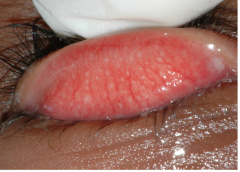
Trachoma Assessment with Eyelid Photographs: Trachoma programs base treatment decisions on the community prevalence of the clinical signs of trachoma, assessed by direct examination of the conjunctiva. Automated assessment could be more standardized and more cost-effective. We tested the hypothesis that an automated algorithm could classify eyelid photographs better than chance. A total of 1,656 field-collected conjunctival images were obtained from clinical trial participants in Niger and Ethiopia. Images were scored for trachomatous inflammation—follicular (TF) and trachomatous inflammation—intense (TI) according to the simplified World Health Organization grading system by expert raters. We developed an automated procedure for image enhancement followed by application of a convolutional neural net classifier for TF and separately for TI. One hundred images were selected for testing TF and TI, and these images were not used for training. The agreement score for TF and TI tasks for the automated algorithm relative to expert graders was κ = 0.44 (95% CI: 0.26 to 0.62, P < 0.001) and κ = 0.69 (95% CI: 0.55 to 0.84, P < 0.001), respectively. For assessing the clinical signs of trachoma, a convolutional neural net performed well above chance when tested against expert consensus. Further improvements in specificity may render this method suitable for field use. |
|
|
Development of the Next-Generation Myoelectric Controlled Prosthetic Arms: There are over 32 million amputees worldwide whose lives are severely impacted by their limb losses. With the advances of electromyography (EMG) signal processing, myoelectric controlled prosthetic arm, which deciphers movement intentions from EMG signals using pattern recognition algorithms to control prosthetic arms, has shown great potential for intuitive prosthesis control. However, the clinical and commercial impact of existing myoelectric controlled prosthetic arms are still limited because natural limb movements are usually continuous and dynamic; however, existing myoelectric control interfaces based on standard single-channel EMG recordings can only provide discrete decisions of static motions due to the lack of meaningful neuromuscular information can be captured. Our research aims to address this challenge and develop the next-generation myoelectric controlled arms by integrating grid sensing, machine learning, and neuromorphic computing technologies. Supported by 2016 Ken Fong Translational Research Fund, SFSU Center for Computing for Life Sciences (CCLS) Mini Grant. News, Publications: |
|
|
MyoHMI: A Low-Cost and Flexible Platform for Developing Real-Time Human Machine Interface for Myoelectric Controlled Applications: Existing research platforms for developing EMG pattern recognition algorithms are typically based on MATLAB and the collection of EMG signals is often done by expensive, non-portable data acquisition systems. The requirement of these resources usually limits the use of these platforms in the lab environments and prohibits their widespread to other fields and applications. To address this limitation, this project aims to develop a low-cost, easy to use, and flexible platform called MyoHMI for developing real-time human machine interfaces for myoelectric controlled applications. MyoHMI facilitates the interface with a commercial EMG-based armband Myo, which costs less than $200 and can be easily worn by the user without the need of special preparation. MyoHMI also provides a highly modular and customizable C/C++ based software engine which seamlessly integrates a variety of interfacing and signal processing modules, from data acquisition through signal processing and pattern recognition, to real-time evaluation and control. Video, Publications: SMC16, ASEEPSW17, EMBC17 |
|
|
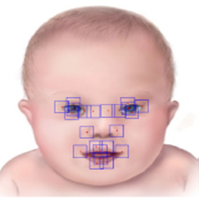
Digital Facial Dysmorphology for Genetic Screening: Summary: To address the lack of safe, reliable, accessible, and economical screening for genetic diseases, such as Down syndrome, in infants after their birth, we have developed a non-invasive protocol that automatically analyzes and classifies infant’s facial images. Techniques such as independent component analysis (ICA) and constrained local models (CLM) are adapted to yield 96% accuracy, superior to the accuracy by human experts , Investigators: Okada (Collaborators, Drs. Zhao & Linguraru, CNMC), Publications: EMBC13, MICCAI13, MIA14, EMBC14, ISBI15, US9443132 |
|
|
Lung Biomarkers for Children: Summary: Viral infection is a leading cause of death for children worldwide. Common diagnostic tests for adults, such as PFT and CT scans, are ineffective for pediatric population due to their inability to follow oral instructions and high radiation risk. We propose a diagnostic measure to quantify severity of lung infection by assessing inhomogeneity of lung fields due to air trapping by analyzing X-ray images of children, Investigators: Golbaz, Okada (Collaborators, Drs. Nino & Linguraru, CNMC), Publications: IJBI13, CARS15, EMBC15, ATSIC16 |
|
|
Protein Crystallization Image Classification with Class-Imbalanced Datasets: Summary: Protein crystallization plays a crucial role in pharmaceutical research by supporting the investigation of a protein’s molecular structure through X-ray diffraction of its crystal. Due to the rare occurrence of crystals, images must be manually inspected, a laborious process. We develop a solution incorporating a regularized, logistic regression model for automatically evaluating these images. Our particular focus is to investigate how best to train a ML classifier in the presence of severe class-skews in datasets. Investigators: Hung, Weldetsion, Newland, Chiang, Collins, Okada (Collaborators, Genentech, Inc.), Publications: SPIE14 |
|
|
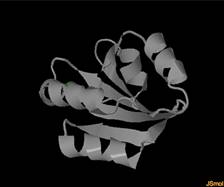
Protein Function Analysis with Microenvironments by Random Forest: Summary: Machine learning-based prediction of protein functions plays a key role in bioinformatics and pharmaceutical research, facilitating swift discovery of new drugs in high-throughput settings. We investigate an adaptation of Random Forest to the structure-based protein function prediction. Our system represents protein’s 3D physicochemical structural information in microenvironment descriptors whose spatial resolution is much finer than other sequence-based protein descriptors. Variable importance measures of RF are also investigated as a tool for mining important genes for particular functional class. Investigators: Okada (Collaborators, Dr. Altman, Stanford, Dr. Petkovic, SFSU), Publications: ICPR14 |
|
|
Variable Interaction Analysis with Random Forest Classifiers: Summary: variable interaction quantifies the extent of complex statistical interaction between a pair of variables beyond linear correlation. We explored such a measure in a supervised classification setting using the well-known Random Forest. We propose 1) an explicit formula for the Gini-based variable interaction originally proposed by Breiman, as well as a novel permutation-based variable interaction extending the well-known permutation-based variable importance measure of RF. Investigators: Hong, Okada, Publications: ISBI12 |
|
|
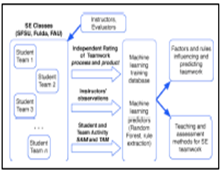
Assessment and Prediction of Teamwork Effectiveness in Software Engineering Courses: Summary: One of the challenges in effective software engineering (SE) education is the lack of objective assessment methods of how well student teams learn the critically needed teamwork practices. We propose a novel machine learning approach to quantitatively assess and predict student learning of teamwork effectiveness in software engineering education based on: a) extracting only objective and quantitative student team activity data during their team class project; b) pairing these data with related independent observations and grading of student team effectiveness in SE process and SE product components. Investigators: Okada, (Collaborators, Dr Petkovic, SFSU, Dr. Huang FAU, Dr. Todtenhoefer, UAS Fulda), Publications: FIE12, ACMSIGSOFT13, FIE14, FIE16 |
|
|
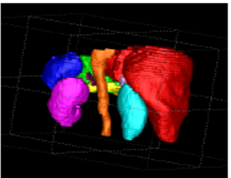
Multi Organ Segmentation with Missing Organs in Abdominal CT Images: Summary: In developing a CAD solution for cancers, our target patient population will have a high likelihood of previous surgical organ resection, however most state-of-the-art organ segmentation methods ignore this fact. We propose a statistical method to detect missing organs and simultaneously segment 10 abdominal organs in CT, extending the atlas-based multi-organ segmentation by Shimizu et al. Investigators: Suzuki, Okada (Collaborators, Dr. Linguraru), Publications: Abdoimg11, MICCAI12 |
|
|
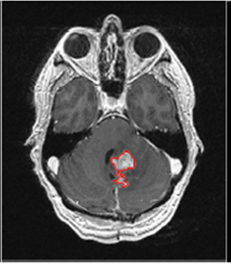
GBM Brain Tumor Segmentation & Classification in Diffusion-Weighted MRI, Summary: Glioblastoma multiforme (GBM) is the most common and most aggressive malignant primary brain tumor in humans. GBM has the worst prognosis of any CNS malignancy. Image-based delineation and diagnosis of GBM remains a significant challenge. We propose a robust Computer-Aided Therapeutic Response (CARrx) paradigm by using the diffusion weighted MRI as a surrogate biomarker to reveal changes in the tumor microenvironment that precedes morphologic tumor changes. To account for the large morphological change of GBM, we propose ensemble segmentation algorithms that combine results from multiple segmentation algorithms as well as perturbations due to user-interaction. Investigators: Huo, Okada, (Collaborator, Dr. Brown, UCLA), Publications: Algorithm09, CDMRI08, SPIE09, SPIE11a, SPIE11b, MLCAD12, MedPhys13, ClinRadiol15 |
|
|
Data/Text Mining for Automatic Lesson Planner for Special Education, Summary: We develop an online web-based tool for assisting teaching credential students in a mild-to-moderate special education program in CA to create sound lesson plans and manage their e-portfolios. Despite tremendous challenges in preparing next generation special education teachers to meet our classroom demands, there are currently no useful tools in practice to assist these teacher candidates to learn how to make sound lesson plans for specific IEPs and regulatory content standards. Our goal is to develop a software tool that automatically suggests some evidence-based teaching strategies that conforms to specific IEP and content-standards through analyzing published peer-reviewed articles by using data mining techniques. Investigators: Sun, Shao, Lam-Miller, (Collaborators, Dr. Courey, SFSU), Publications: CSEDU09, SITE10, EDMEDIA10 |
|
|
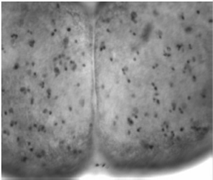
Biological 2D Cellular Image Analysis, Summary: This project aims to develop suits of tools to analyze technically challenging biomedical images, such as 2D optical images of immunostained cell nuclei in nervous systems of Tobacco hornworm for studying the effect of starvation in neural development. Our cell counting solution for the Tobacco hornworm images exploits fast radial symmetry transform originally proposed for generic object/face detection. Investigators: Timilsina, (Collaborators, Dr. Moffatt, SFSU), Publications: SPIE11 |
|
|
3D Topology Type Classification in 3D Chest CT, Summary: Correct classification of topological types of local anatomical and pathological structures plays a crucial role in many 3D medical image analysis applications, such as vascular and airway tree analysis, detection of internal bleeding, and false-positive detection for lung nodule detection. We propose a novel algorithm to classify a local 3D structure into four types of topology in 3D (i.e., blob, plane, tube, and branching-tube). The key idea is to transform the analysis of 3D objects into a 2D clustering problem which can be solved efficiently. The proposed algorithm is also extended to Bayesian formalism with a novel mean shift clustering method that accounts for our circular domain data. Investigators: Miao, Kafai, Publications: SPIE06, MLMI10, MMBIA10 |
|
|
Directional Mean Shift, Summary: Mean shift is a convergent hill-climbing algorithm that efficiently seeks a mode of kernel-smoothed function such as kernel density functions and Gaussian-smoothed data. Mean shift has been successfully applied to various vision applications, such as clustering, segmentation, tracking to name a few. Our research extends this mean shift to a wider range of data analysis tasks, including Gaussian scale space, MAP estimation, and directional/circular domain data analysis. Investigators: Kafai, Okada, Publications: MMBIA10 |
|
|
Dental Lesion CAD in 3D Cone Beam CT, Summary: In endodontics, 3D cone beam CT has shown a promise for classifying granulomas (tissue infection) and cysts, which could not be differentiated by traditional X-ray imaging in past, leading to potentially unnecessary surgical removals of these lesions. We propose a new CAD scheme that offers a non-invasive differential diagnosis of the two types of dental lesions to avoid unnecessary tissue damage. Our solutions combine graph-based random walker segmentation and a new LDA-Boost algorithm for diagnostic classification. Investigators: Rysavy, Flores, (Collaborators, Dr. Enciso, USC), Publications: SPIE08, ICPR08, ISBI09, MLMI10, MedPhys15 |
|
|
Lung Nodule Analysis in 3D Chest CT, Summary: Lung cancer accounts for the most cancer death in the US. These cancers often appear as focal concentration of high intensity in CT scans, known as lung nodules. This project aims to provide comprehensive suits of robust and accurate detection, segmentation, and analysis of such nodules for early detection and accurate staging. In particular, our method can robustly segment technically challenging Ground-Glass Opacity (GGO) nodules which pose higher malignancy rate than more common solitary nodules. (Example Results: MICCAI04). Investigators: Okada, Publications: TMI05, ECCV04, CVPR04, CVPR05, MICCAI04, MICCAI05, CVBIA05, KDD06, M3ISR10, CADLI10 |
|
|
Cross-Bin Histogram Distance Measures: EMD_L1 & Diffusion Distance, Summary: Histogram features, such as SIFT and Shape Context, are ubiquitous tools in computer vision and pattern recognition. Earth mover’s distance is a well-known cross-bin metric of such histogram features accounting for ill-aligned histogram bins; however it suffers from high computational cost that is larger than O(n^3). We propose an O(n^2) exact earth mover’s distance algorithm. We simplify the underlying linear program by exploiting the L1 ground distance. We further improve the computation complexity in cross-bin distance computation, proposing Diffusion Distance whose cost is reduced to near O(n), while maintaining high scores on SIFT and shape matching tasks we evaluated. Investigators: Ling, Okada, Publications: PAMI06, CVPR06 |
2021:
- T. Touati*, K. Okada, I. Song, What makes a person obese?: An individual-level analysis of obesity, Accepted as 1-page extended abstract at IEEE International Conference on Biomedical and Health Informatics, (2021)
2020:
- G. Urban, S. Porhemmat*, M. Stark*, B. Feeley, K. Okada, P. Baldi, Classifying Shoulder Implants in X-ray Images using Deep Learning, Computational Structural Biotechnology Journal, 18: 967-972 (2020)
2019:
- M. Kim*, K. Okada, A. Ryner, A. Amza, Z. Tadesse, S. Cotter, B. Gaynor, J. Keenan, T. Lietman, T. Porco, Sensitivity and specificity of computer vision classification of eyelid photographs for programmatic trachoma assessment, PLoS One, 14(2): e0210463 (2019)
- A. Mansoor, J. Cerrolaza, G. Perez, E. Biggs, K. Okada, G. Nino, M. Linguraru, A Generic Approach to Lung Field Segmentation from Chest Radiographs using Deep Space and Shape Learning, IEEE trans Biomedical Eng., Epub: doi: 10.1109/TBME.2019.2933508 (2019)
2018:
- R. Khilari*, J. Puchin*, K. Okada, Automated quasi-3D spine curvature quantification and classification, In proc. SPIE Medical Imaging, Huston, 2018
- J. Puchin*, X. Zhang, K. Okada, Independent Component Analysis for Hand Gesture Recognition with EMG Sensor Grid, Submitted to IEEE EMBC, Honolulu, 2018
- I. Donovan*, K. Okada, X. Zhang, Adjacent Feature for High-Density EMG Pattern Recognition, Submitted to IEEE EMBC, Honolulu, 2018
- A. Kulkarni, I. Yoon, K. Okada, P. Pennings, C. Domingo, Promoting Diversity in Computing, Submitted to ACM ITiCSE, Lanarca, Cyprus, 2018
- I. Yoon, K. Okada, A. Kulkarni, P. Pennings, C. Domingo, Promoting Inclusivity in Computing (PINC) via Computing Application Minor, Accepted the American Society for Engineering Education 2018 (ASEE), 2018
2017:
- I. Donovan*, J. Puchin*, K. Okada, X. Zhang, Simple Space-Domain Features for Low-Resolution sEMG Pattern Recognition, In proc. IEEE Eng. Med. Biol. Soc. (EMBC), Korea, 2017 PDF
- J. Yan*, J. Dalton*, B. Doronila*, K. Chang-Kam*, V. Melara*, C. Thomas*, I. Donovan**, K. Bholla**, A. G. Enriquez, W. Pong, Z. Jiang, C. Chen, K.-S. Teh, H. Mahmoodi, H. Jiang, K. Okada, and X. Zhang, Engaging Community College Students in Computer Engineering Research through Design and Implementation of a Versatile Gesture Control Interface, In proc. the American Society for Engineering Education 2017 Pacific Southwest Conference (ASEE/PSW-2017), Tempe, AZ, April 20-22, 2017. PDF
2016:
- I. Donovan*, K. Valenzuela*, A. Ortiz*, S. Dusheko*, H. Jing, K. Okada, X. Zhang, MyoHMI: A Low-Cost and Flexible Platform for Developing Real-Time Human Machine Interface for Myoelectric Controlled Applications, In proc. IEEE Int Conf System, Man and Cybernetics (SMC), 2016 PDF
- D. Petkovic, M. Sosnick-Perez, K. Okada, R. Todtenhoefer, N. Miglani*, A. Vigil*, Using the Random Forest Classifier to Assess and Predict Student Learning of Software Engineering Teamwork, In proc. Frontiers in Education Conference (FIE), 2016 PDF
- A. Mansoor, G. F. Perez, K. Pancham, A. Jain, K. Okada, M.G. Linguraru, G. R. Nino, Automated Lung Segmentation and Longitudinal Air Trapping Quantification in Chronic Lung Disease of Prematurity, In proc. American Thoracic Society International Conference, pp A4575, 2016 LINK
- M.G. Linguraru, Q. Zhao, K. Rosenbaum, M. Summar, K. Okada, Device and Method for Classifying a condition based on Image Analysis, US Patent, US9443132 PDF
2015:
- K. Okada, S. Rysavy*, A. Flores*, MG. Linguraru, Noninvasive differential diagnosis of dental periapical lesions in cone-beam CT scans, Medical Physics, 42(4): 1653-1665 (2015) LINK
- J. Huo*, J. Alger, H Kim, M Brown, K. Okada, W. Pope, J. Goldin, Between-Scanner and Between-Visit Variation in Normal White Matter Apparent Diffusion Coefficient Values in the Setting of a Multi-Center Clinical Trial, Clinical Neuroradiology, EPub: March 20 (2015) LINK
- Q. Zhao*, K. Okada, K. Rosenbaum, M. Summar, MG. Linguraru, Constrained Local Model with Independent Component Analysis with Kernel Density Estimation: Application Down syndrome Detection, In proc. IEEE Int. Symp. Biomed. Imaging (ISBI), 967-970, New York, 2015 PDF
- A. Mansoor, GF. Perez, K. Pancham, A. Khan, K. Okada, G. Nino, MG. Linguraru, Partitioned Active Shape Model With Weighted Landmarks For Accurate Lung Field Segmentation, In proc. Computer-Assisted Radiology and Surgery (CARS) Extended Abstract, S224-6, Barcelona, 2015 PDF
- K. Okada, M. Golbaz*, A. Mansoor, GF. Perez, K. Pancham, A. Khan, G. Nino, MG. Linguraru, Severity Quantification of Pediatric Viral Respiratory Illnesses in Chest X-Ray Images, In proc. IEEE Eng. Med. Biol. Soc. (EMBC), Invited Paper, Milan, 2015 PDF
2014:
- Q. Zhao, K. Okada, K. Rosenbaum, L. Kehoe, DJ. Zand, R. Sze, M. Summar, MG. Linguraru, Digital facial dysmorphology for genetic screening: Hierarchical constrained local model using ICA, Medical Image Analysis, 18(5): 699-710 (2014) LINK
- CS. Mendoza, N. Safdar, K. Okada, E. Myers, GF. Rogers, MG. Linguraru, Personalized assessment of craniosynostosis via statistical shape modeling, Medical Image Analysis, 18(4): 635-46 (2014) LINK
- EA. Simpson, KV, Jakobsen, DM. Fragaszy, K. Okada, JE. Frick, The development of facial identity discrimination through learned attention, Developmental Psychobiology, 56(5): 1083-1101 (2014) LINK
- Q. Zhao*, N. Werghi, K. Okada, K. Rosenbaum, M. Summar, MG. Linguraru, Ensemble Learning for the Detection of Facial Dysmorphology, In proc. IEEE Eng. Med. Biol. Soc. (EMBC), 2014:3633-636, Chicago, 2014 PDF
- D. Petkovic, M. Sosnick-Perez, , S. Huang, R. Todtenhoefer, K. Okada, S. Arora*, R. Sreenivasen*, L. Flores*, S. Dubey*, SETAP: Software Engineering Teamwork Assessment and Prediction Using Machine Learning, In proc. Frontiers in Education Conference (FIE), 2014 PDF
- J. Hung*, J. Collins*, M. Weldetsion*, O. Newland*, E. Chiang*, K. Okada, Protein crystallization image classification with elastic net, In proc. SPIE Medical Imaging, San Diego, 2014 PDF
- K. Okada, L. Flores*, M. Wong, D. Petkovic, Microenvironment-Based Protein Function Analysis by Random Forest, In Proc. International Conference on Pattern Recognition, Stockholm, 2014 PDF
2013:
- J. Huo*, K. Okada, EM. van Rikxoort, HJ Kim, JR Alger, WB Pope, JG Goldin, MS Brown, Ensemble segmentation for GBM brain tumors on MR images using confidence-based averaging, Medical Physics, 40(9): 093502 (2013) PDF
- S. Huang, D. Petkovic, K. Okada, M. Sosnick, S. Zhu, R. Todtenhoefer, Toward objective and quantitative assessment and prediction of team work effectiveness in software engineering courses, ACM SIGSOFT Software Engineering Notes, 38(1): 7-9 (2013) PDF
- A. El-Baz, GM. Beache, GL. Gimel’farb, K. Suzuki, K. Okada, A. Elnakib, A. Soliman, B. Abdollahi, Computer-aided diagnosis systems for lung cancer: challenges and methodologies, International Journal of Biomedical Imaging, 2013:942353 (2013) PDF
- A. El-Baz, GM. Beache, GL. Gimel’farb, K. Suzuki, K. Okada, Editorial: Lung Imaging Data Analysis, International Journal of Biomedical Imaging, 2013:618561 (2013) LINK
- Q. Zhao*, K. Okada, K. Rosenbaum, DJ. Zand, R. Sze, M. Summar, MG. Linguraru, Hierarchical Constrained Local model Using ICA and Its Application to Down Syndrome Detection, In Proc. Int. Conf. Medical Image Computing and Computer Assisted Intervention (MICCAI), vol.2, pp. 222-229, Nagoya, 2013 PDF
- Q. Zhao*, K. Rosenbaum, K. Okada, DJ. Zand, R. Sze, M. Summar, MG. Linguraru, Automated down syndrome detection using facial photographs, In proc. IEEE Eng. Med. Biol. Soc. (EMBC), 2013:3670-3, Osaka, 2013 PDF
- J. Collins*, K. Okada, Learning metrics for content-based medical image retrieval, In proc. IEEE Eng. Med. Biol. Soc. (EMBC), 2013:3363-6, Osaka, 2013 PDF
2012:
- J. Huo*, M.S. Brown, K. Okada, CADrx for GBM Brain Tumors: Predicting Treatment Responses from Changes in Diffusion-Weighted MRI, In Machine Learning in Computer-Aided Diagnosis: Medical Imaging Intelligence and Analysis, IGI-Global, 2012 PDF
- M. Suzuki*, M.G. Linguraru, K. Okada, Multi-Organ Segmentation with Missing Organs in Abdominal CT Images, Accepted, Int. Conf. Medical Image Computing and Computer Assisted Intervention (MICCAI), 2012 PDF
- D. Petkovic, K. Okada, M. Sosnick*, A. Iyer*, S. Zhu*, R. Todtenhoefer, S. Huang, A Machine Learning Approach for Assessment and Prediction of Teamwork Effectiveness in Software Engineering Education, Accepted, Frontiers in Education Conference (FIE), 2012 PDF
- C. Kelly*, K. Okada, Variable Interaction Measures with Random Forest Classifiers, In proc. IEEE Symposium on Biomedical Imaging (ISBI), 2012 PDF
- S. Sergeev*, MG. Linguraru, K. Okada, Medical image registration using machine learning-based interest point detector, In proc. SPIE Medical Imaging, 2012 PDF
- J. Huo*, K. Okada, W. Pope, M. Brown, Improving semi-automated segmentation by integrating learning by active sampling , In proc. SPIE Medical Imaging, 2012 PDF
- J. Collins*, K. Okada, A Comparative Study of Similarity Measures for Content-Based Medical Image Retrieval, In CLEF Online Working Notes, 2012 PDF
2011:
- K. Okada, Anisotropic Scale Selection, Robust Gaussian Fitting, and Pulmonary Nodule Segmentation in Chest CT Scans, In Multi-Modality State-Of-The-Art Medical Image Segmentation and Registration Methodologies, Springer, 2011 PDF
- K. Okada, Ground-Glass Nodule Characterization in High-Resolution CT Scans, In Computer-Aided Diagnosis of Lung Imaging, Taylor and Francis, LLC, 2011 PDF
- M. Suzuki*, M.G. Linguraru, R.M. Summers, K. Okada, Analyses of Missing Organs in Abdominal Multi-Organ Segmentation, In proc. MICCAI 2011 Workshop on Computational and Clinical Applications in Abdominal Imaging, 2011 PDF
- N. Timilsina*, C. Moffatt, K. Okada, Development of a Stained Cell Nuclei Counting System, In proc. SPIE Medical Imaging, 2011 PDF
- J. Huo*, K. Okada, W. Pope, M. Brown, Sampling-Based Ensemble Segmentation against Inter-Operator variability, In proc. SPIE Medical Imaging, 2011 PDF
- J. Huo*, E. van Rikxoort, K. Okada, H. Kim, W. Pope, J. Goldin, M. Brown, Confidence-based Ensemble for GBM brain tumor segmentation, In proc. SPIE Medical Imaging 2011 PDF
2010:
- K. Okada, A. Flores*, M. G. Linguraru, Boosting Weighted Linear Discriminant Analysis, International Journal of Advanced Statistics for Economics and Life Sciences, 2(1): 1-11 (2010)
- K. Okada, Pose-Invariant Face recognition: Analysis, Synthesis, Identification of Human Faces with Pose Variations, VDM Verlag Dr. Mueller, 2010
- K. Okada, Mean Shift from Theory to Application, In Computer Vision Saisentan Guide 2, Adcom Media, 2010 PDF
- Y. Miao*, M. Kafai*, K. Okada, Bayesian Classification of Local 3D Structures in Medical Images, In proc. International Workshop on Machine Learning in Medical Imaging (MLMI), 2010 PDF
- A. Flores*, M. G. Linguraru, K. Okada, Boosted-LDA for Biomedical Data Analysis, In proc. International Workshop on Machine Learning in Medical Imaging (MLMI), 2010 PDF
- M. Kafai*, Y. Miao*, K. Okada, Directional Mean Shift and its Application for Topology Classification of Local 3D Structures, In proc. IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis (MMBIA), 2010 PDF
- S. Courey, X. Shao*, N. Lam-Miller*, K. Okada, Web-Based Tools for Enhancing Teacher Preparation: Helping to Build a High Quality Teaching Workforce, In proc. World Conference on Educational Multimedia, Hypermedia & Telecommunications (ED-MEDIA), 2010 PDF
- X. Shao*, N. Lam-Miller*, S. Courey, K. Okada, A Web-based Lesson Plan Creator for Teaching Preparation Programs, In proc. Annual Conference for Society for Information Technology & Teacher Education (SITE), 2010 PDF
2009:
- J. Huo*, K. Okada, H. Kim, W. Pope, J. Goldin, J. Alger, M. Brown, CADrx for GBM Brain Tumors: Predicting Treatment Response from Changes in Diffusion-Weighted MRI, Algorithms, 2: 1350 – 1367 (2009) PDF
- A. Flores*, S. Rysavy*, R. Enciso, K. Okada, Non-Invasive Differential Diagnosis of Dental Periapical Lesions in Cone-Beam CT, In proc. IEEE International Symposium of Biomedical Imaging: From Nano to Macro (ISBI), 2009 PDF
- K. Okada, N. Lam-Miller*, X. Shao*, S. Courey, Web-based Tools for Credential Candidates in Special Education Programs, In proc. International Conference on Computer Supported Education (CSEDU), 2009 PDF
- J. Huo*, W. Pope, G. Kim, K. Okada, M. Brown, J. Alger, Y. Wang, J. Goldin, Histogram-based classification with Gaussian Mixture Modeling for GBM Tumor Treatment Response using ADC Map, In proc. SPIE Medical Imaging Conferences, Orland, 2009 PDF
2008:
- K. Okada and C. von der Malsburg, Face Recognition and Pose Estimation with Parametric Linear Subspaces, In Applied Pattern Recognition, H. Bunke, A. Kandel, M. Last (Eds.), pp. 49-74, Springer, 2008 PDF
- S. Rysavy*, A. Flores*, R. Enciso, K. Okada, Segmentation of Large Periapical Lesions toward Dental Computer-Aided Diagnosis in Cone-Beam CT Scans, In proc. SPIE Medical Imaging Conferences, vol. 6914, pp. 153, San Diego, 2008 [Best poster honorable mention award] PDF
- S. Rysavy*, A. Flores*, R. Enciso, K. Okada, Classifiability Criteria for Refining of Random Walks Segmentation, In proc. International Conference on Pattern Recognition (ICPR), Tampa, 2008 PDF
- J. Huo*, W. Pope, K. Okada, J. Alger, HJ. Kim, Y. Wang, J. Goldin, M. Brown, Early Detection of Treatment Response for GBM Brain Tumor using ADC Map of DW-MRI, In proc. MICCAI 2008 Workshop on Computational Diffusion MRI, New York, 2008 PDF
- K. Okada, S. Periaswamy, J. Bi, Stratified Regularity Measures with Jensen-Shannon Divergence, In proc. IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis (MMBIA), Anchorage, 2008 PDF
- T. Goh*, R. West*, K. Okada, Robust Detection of Semantically Equivalent Visually Dissimilar Objects, In proc. International Workshop on Semantic Learning Applications in Multimedia (SLAM), Anchorage, 2008 PDF
2007:
- H. Ling* and K. Okada, An Efficient Earth Mover’s Distance Algorithm for Robust Histogram Comparison, IEEE Trans. Pattern Analysis and Machine Intelligence, 29(5): 840-863 (2007) PDF
- J. Martin-Malivel and K. Okada, Human and Chimpanzee Face Recognition in Chimpanzees (Pan troglodytes): Role of Exposure and Impact on Categorical Perception, Behavioral Neuroscience, 121(6): 1145-1154 (2007) PDF
- K. Okada and X. Huang, Robust Click-Point Linking: Matching Visually Dissimilar Local Regions, In proc. IEEE International Workshop on Beyond Multiview Geometry: Robust Estimation and Organization of Shapes form Multiple Cue, Minneapolis, 2007 PDF
2006:
- K. Okada, X. Huang, X. Zhou, A. Krishnan, Robust Click-Point Linking for Longitudinal Follow-Up Studies, In proc. Int. Workshop on Medical Imaging and Augmented Reality (MIAR), pp. 252-260, Shanghai, 2006 PDF
- K. Okada, M. Singh, V. Ramesh, A. Krishnan, Prior-Constrained Scale-Space Mean Shift, In proc. British Machine Vision Conference (BMVC), vol. II, pp. 829-838, Edinburgh, 2006 PDF
- J. Bi, S. Periaswamy, K. Okada, T. Kubota, G. Fung, M. Salganicoff, B. Rao, Computer Aided Detection via Asymmetric Cascade of Sparse Hyperplane Classifiers, In proc. Annual SIGKDD Int. Conf. on Knowledge Discovery and Data Mining (KDD), pp. 837-844, Philadelphia, 2006 PDF
- H. Ling* and K. Okada, Diffusion Distance for Histogram Comparison, In proc. IEEE Conf. on Computer Vision and Pattern Recognition (CVPR), vol. I, pp. 246-253, New York, 2006 PDF
- H. Ling* and K. Okada, EMD_L1: An Efficient and Robust Algorithm for Comparing Histogram-Based Descriptors, In proc. European Conf. Computer Vision (ECCV), vol. III, pp. 330-343, Graz, 2006 PDF
- C. Bahlmann, X. Li*, K. Okada, Local Pulmonary Structure Classification for Computer-aided Nodule Detection, In proc. SPIE Medical Imaging Conferences, vol. 6144, pp. 1775-1785, San Diego, 2006 PDF
Kazunori Okada, Ph.D.
kazokada AT sfsu.edu
https://bidal.sfsu.edu/~kazokada/
Computer Science Department
San Francisco State University
1600 Holloway Ave. San Francisco, CA 94132
Currently no positions are open.
© Kazunori Okada, 2011, 2012, 2013, 2014, 2015, 2016, 2017, 2018, 2019, 2020, 2021, 2022, 2023
San Francisco State University, San Francisco CA, USA